When you log onto LinkedIn, you’re normally presented with suggestions to connect with people you know, either because you went to the same university as them, or worked in the same company or industry.
However, the suggestions can sometimes take us by surprise, like when the algorithm recommends a relative or family friend even though they work in a totally different field to you. Given the total lack of professional overlap, you might wonder how LinkedIn could possibly know about these real life relationships.
The artificial intelligence (AI) algorithms that drive these recommendations use a type of technology known as a Graph Neural Network, which is based on graphs: mathematical structures made up of nodes and the links (also known as “edges”) that connect them. For a social network such as LinkedIn, a graph can be generated where the nodes represent each user, while the links are the connections between them.
These algorithms collect information from the immediate environment of each node – our direct connections on LinkedIn. They then aggregate that information and integrate it into the original node.
After this process, each profile reflects both its own data and that of its immediate network. This process can be carried out several times – in the second iteration, when we aggregate information from our neighbours, they in turn will already have aggregated information from their own neighbours and, consequently, we will have information from the second neighbourhood.
M. Hernaez / BioRender.
A web of relationships
In these networks, it is not just our own personal information that matters, but also who we have connected with and who our connections have connected with. In the full version of LinkedIn’s algorithm, as used in practice, there are not only nodes representing people, but also other types of nodes, such as companies or publications.
This means the algorithm can get information both from our personal connections and from the content we have marked as favourites or interacted with.
If, for instance, someone has your sister as a connection and has “liked” posts that your brother-in-law also likes, the algorithm can detect that you not only share similar interests, but that you may also be personally connected in some way.
Social media algorithms in biomedicine
Developing a drug from scratch is extremely costly and time-consuming. The discovery process often resembles a funnel. At the top, all potential candidates enter and, after being narrowed down through various stages of research, only one is left to enter clinical trials. This drug will then (hopefully) pass through to become available for clinical use among the general population.
Though it is necessary, the complexity of this provess means drug repurposing has become increasingly common in recent decades. The aim of this process is not to design new drugs, but to find new uses for existing ones.
To treat a disease, we generally focus on targeting the proteins responsible for it. There are public and well-documented databases containing information on which proteins each drug targets, and these databases have grown considerably in recent years.
One of the most widely used databases, DrugBank, has gone from 841 approved drugs when it was first released in 2006, to 2,751 in its most recent 2024 update. This growing availability of data allows for the use of more complex models.
With this volume of data, we can create a graph network where the nodes are drugs and proteins, and the links are the interactions between them, as recorded in databases. Once we have the network, we can then apply similar algorithms to those used in social media: for each drug, we add biochemical information about the proteins with which it interacts through the known connections.
Using this information, the model can then tell us the probability of a drug-protein interaction that we did not previously have in the database, as the algorithms can efficiently analyse large volumes of information. These interactions can then be validated under laboratory conditions, saving time and money from the lengthy discovery process.
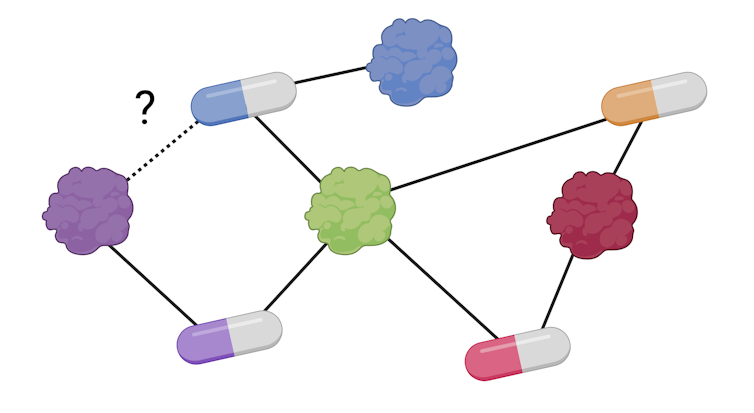
M. Hernáez / BioRender
Our research
At the Computational Biology and Translational Genomics lab at the University of Navarra, we have followed this idea to develop GeNNius, a model that aims to build a network between drugs and proteins. Its implementation has already improved existing models, especially in terms of run time: in just one minute we can evaluate around 23,000 interactions.
While the model has good predictive capabilities, there is still room for improvement. For instance, challenges arise when assessing possible interactions with molecules that are not part of the network, or for which we have little original data. Although it is technically possible to generate an output, the model often gives low confidence results in these cases.
By overcoming these obstacles and with further research, these models could evolve in the future into systems that provide personalised recommendations for each patient.



